Molecular simulation and its application
Molecular simulation is a method to calculate the movement of atoms based on interatomic forces and applied to many situations. Here we have chosen molecular dynamics (MD) which considers the presence of a solvent and have conducted a variety of research including the analysis of a protein-ligand binding process; the analysis of a folding process in which proteins are folded into specific structures in vivo; a study to improve prediction accuracy by modifying structures obtained through simulation which started from a predicted structure generated in a structure prediction program; and the functional analysis of protein molecules. These researches are supervised by Associate Professor Tohru Terada.
- Analysis of protein-ligand binding processes
- Folding simulation
- Analysis of protein functions
- Parallel computing of molecular dynamics simulation
Analysis of protein-ligand binding processes
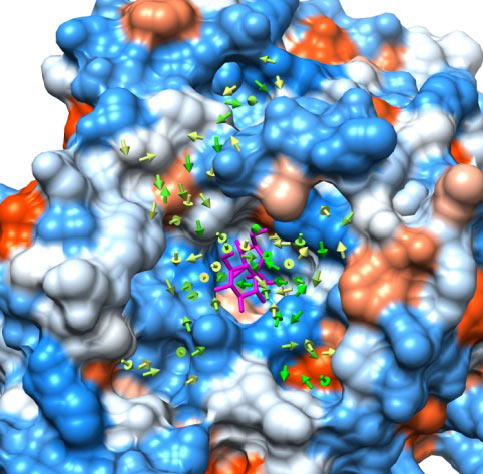
We conduct research which is involved in the analysis of protein-ligand binding processes by molecular simulation using a coarse grained model called MARTINI. To elucidate a ligand binding pathway, ligand movement was analyzed from trajectories that are involved in ligand binding. Several typical ligand and pocket patterns are analyzed, and the figure is the result of analysis which demonstrates a binding event between sucrose and levansucrase. As you can see, the ligand moves on the protein surface toward the binding site.
Folding simulation
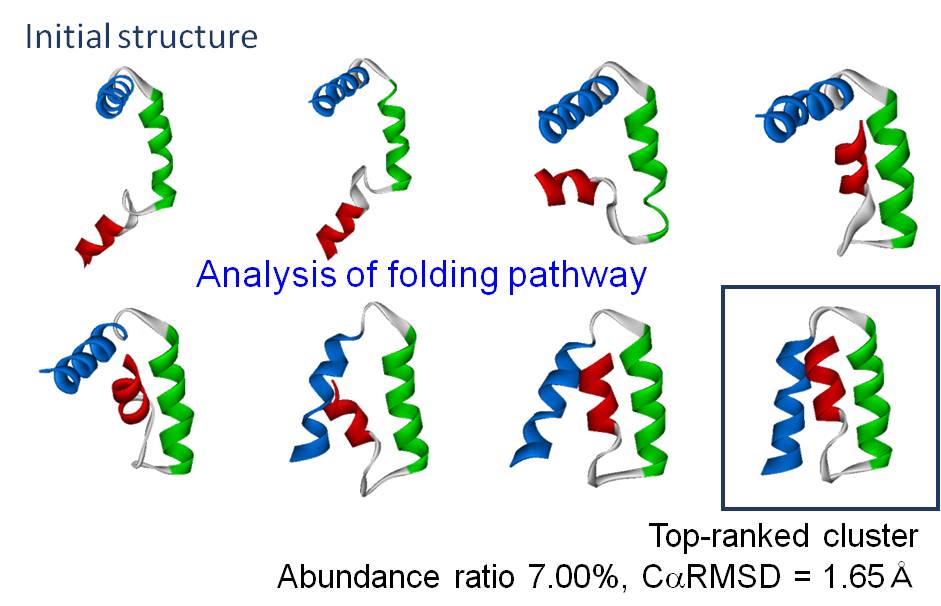
Although the reproduction of a protein folding process requires enormous calculation time, we have successfully demonstrated the folding of various proteins such as chignolin, B domain of protein A, WW domain, and FSD-EY, using efficient structure sampling and methods which calculate the solvent effect or constraint secondary structures. As for chignolin, the folding pathway was analyzed in more detail by conducting mutation experiments and nuclear magnetic resonance (NMR) measurements.
Analysis of protein functions
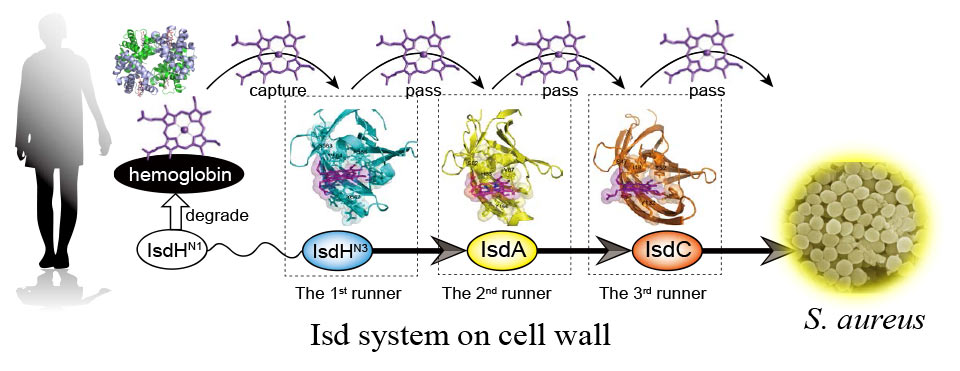
Heme-binding mechanism of structurally similar Isd NEAT domains of Staphylococcus aureus
IsdH-N3, IsdA, and IsdC are studied as the targets of Staphylococcus aureus containment. Although no major structural difference exists among these proteins, heme is transferred in the order of IsdH-N3 → IsdA → IsdC and they display different heme affinities. By combining calculations and experiments, we attempt to uncover the reason for such affinity differences based on protein structures. This research is the collaboration with Professor Kouhei Tsumoto at the University of Tokyo.
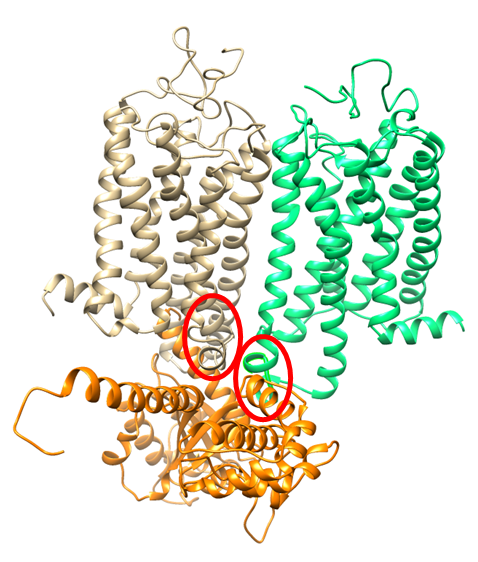
Overdominance effect of the bovine ghrelin receptor (GHSR1a)-DelR242 locus on growth in Japanese Shorthorn weaner bulls
Ghrelin and the ghrelin receptor (GHSR1a) are involved in growth hormone secretion, food intake, and several other important functions. Ghrelin acts on GHSR1a and induces signal transduction via the Gaq subunit. Dr. Masanori Komatsu’s group at NARO Institute of Livestock and Grassland Science of National Agriculture and Food Research Organization (NARO) found that 4R/3R heterozygotes of GHSR have a selective advantage in weaner bulls because of their higher average daily gain than homozygotes. We analyzed possible molecular mechanisms involved in the overdominance effect of the DelR242 locus on these traits in weaner bulls using a structural model of the complex consisting of a GHSR1a dimer and Gaq.
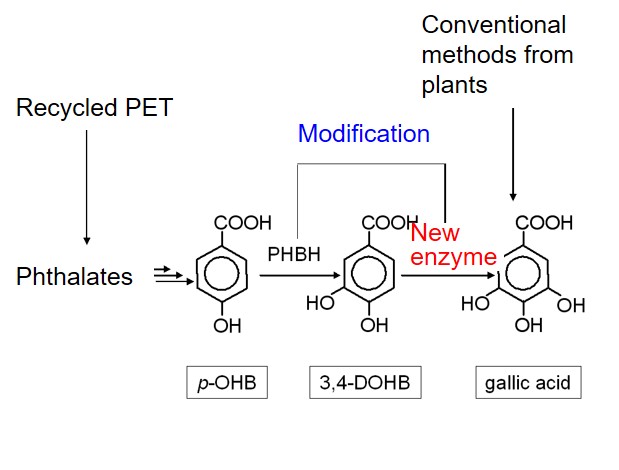
Development of an effective production method of gallic acid by enzyme modification
Gallic acid is mainly used as an important substrate for the synthesis of propyl gallate. It is used as an antioxidant food additive and its alkaline solution is used as a reducing agent. In this research, we employed protein structure modeling and interaction analysis to propose the candidate mutants of p-hydoroxybenzoate hydroxylase(PHBH) which converts 3,4-Dihydroxybenzoate(DOHB) to gallic acid in high yield at a low cost. This research is the collaboration with Genaris, Inc.
Parallel computing of molecular dynamics simulation
We also study the parallel computation of MD using a many-core system.